Development of fluorogen activating proteins (FAPs) over the past two decades has led to the diverse set of over 160 publications from CMU and others as listed below. The unique capabilities of FAPs have facilitated a broad variety of in vivo studies in systems ranging from bacterial, yeast and mammalian cells to nematode, fly, zebrafish and mouse. The following examples illustrate key properties of FAPs that make them powerful tools for basic and biomedical research and drug discovery.
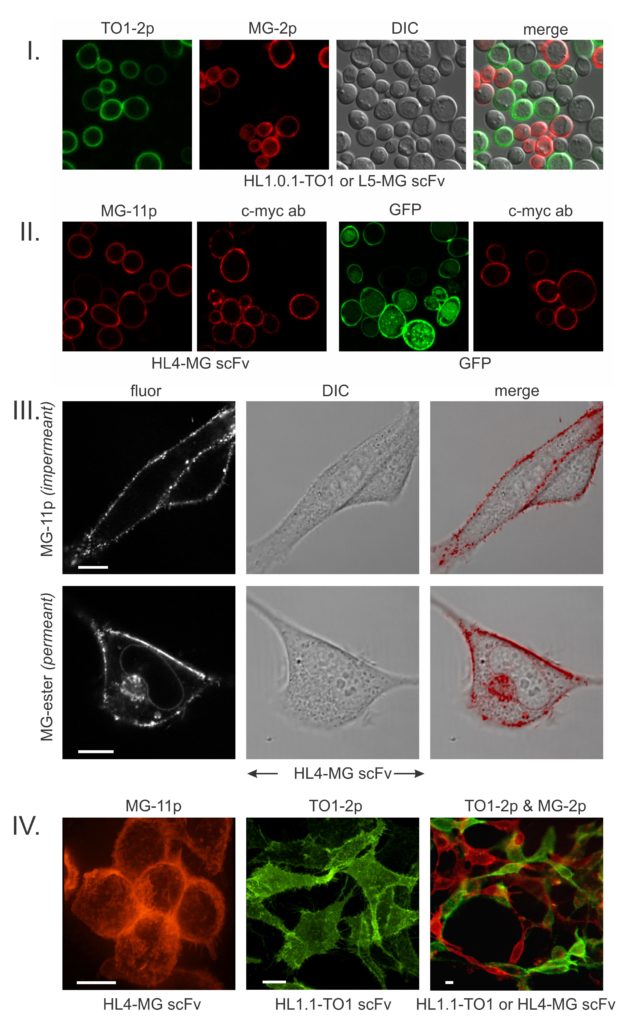
The above images from our seminal paper (ref. 1) establish the basic behavior of FAPs. FAPs were discovered by screening a library of yeast displayed scFvs (antibody fragments) against pegylated fluorogenic dyes (I). The salient characteristic of FAPs that bind pegylated fluorogens is that they generate intense surface-specific fluorescence that for our most useful fluorogens (malachite green, MG-2p; thiazole orange, TO1-2p) are respectively enhanced by thousands-fold. This behavior enables essentially instant fluorescence visualization of a surface-expressed FAP-fusion protein simply by adding a fluorogen to the cell culture without need of washing out excess dye. For cell surface-specific applications, FAPs have a marked advantage over fluorescent proteins (GFPs) which are compromised by internal signal generated by the biosynthetic/secretory pathway (II). Importantly, FAP technology eliminates the need for resource-intensive labeling with dye-conjugated antibodies (II).
If one substitutes the PEG on malachite green with an ester, the fluorogen becomes cell-permeant (III). In these mammalian cells, the permeant fluorogen additionally lights-up the FAP on nuclear envelope ribosomes, Golgi and various secretory vesicles. Intracellular labeling has been used in several studies of nuclear, mitochondrial and Golgi proteins. Inside/outside differential labeling has been especially useful for dynamically tracking the endocytosis/exocytosis of GPCRs and the CFTR ion channel in the context of drug screening and studying the mouse synaptic BK channel. Surface-specific FAP imaging is also useful for visualizing cell/cell contacts (IV).
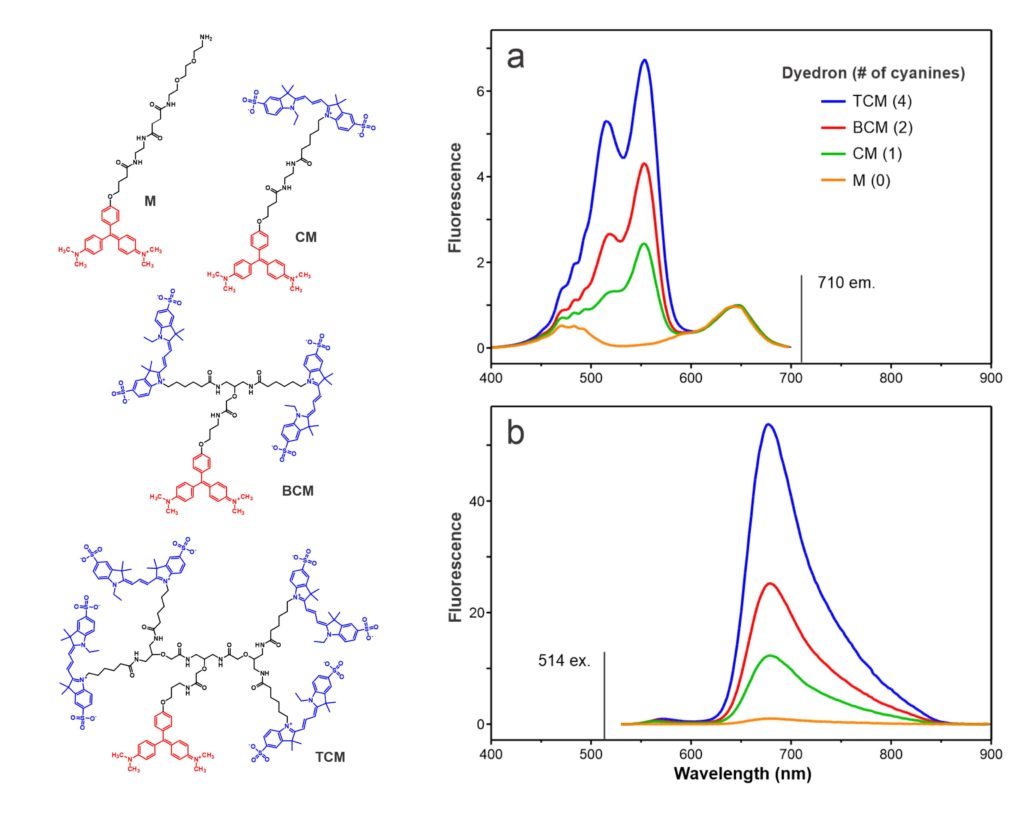
The above chemical structures and fluorescence spectra (ref. 13) depict our first demonstration of the chemical modularity of FAP technology. FAP sensor functionality can readily be modified by conjugating different fluorescent dyes to a fluorogen via the (branched) linker. In this case, a series of Cy3 donors quantitatively amplify the fluorogenic emission of a MG FAP via fluorescence resonance energy transfer (FRET). pH sensitive Cy3 and Cy5 analogs respectively convert MG and TO1 FAPs into sensors that track internalization of cell surface receptors. Other two color bichromophore constructs create spectrally shifted FAPs that can be used for pulse-labeling. MG and TO1 chromophores separated by a defined linker length can be used as a molecular ruler to assess distances between contacting cells respectively displaying cognate FAPs.
FAPs have been used to fluorescently label IgG Fc domains by photochemical conjugation or by using FAPs fused to protein A/G. These FAP/IgG reagents possess useful binding specificity for labeling purposes. Following the chemical modularity paradigm, Biocognon seeks to extend the use of antibody-based reagents by discovering Nanobodies that when fused to FAPs can be used to specifically deliver toxin or radionuclide payloads that are coupled to PEG-fluorogen.
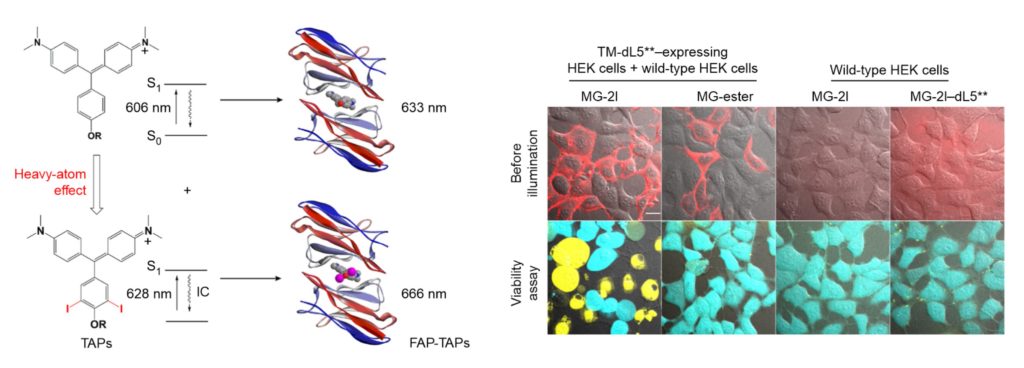
The above fluorogen/FAP schematic and associated cell images document our remarkable discovery that the MG fluorogen can be converted into an efficient FAP-dependent photoablation reagent (ref. 66). When this iodine-modified fluorogen becomes encapsulated within the cognate FAP, it generates cytotoxic reactive oxygen species (ROS) that rapidly kill the host cell. Cell-targeted cytoxicity is entirely dependent on addition of the modified fluorogen, and thus applications are not compromised by background cytoxicity, as is the case with ROS-generating fluorescent proteins. The MG-2I/FAP has been used in transgenic zebrafish to ablate cardiac cells at defined stages of fish development and to ablate mitochondria in cells to study the effects of oxidative stress on telomeres.
Tight encapsulation of unmodified MG by the FAP renders the fluorogen more resistant to photobleaching than fluorescent proteins or fluorescent dyes conjugated to proteins. Photobleaching resistance, together with its far-red emission at 664 nm, makes the MG/FAP complex an exquisitely sensitive reporter for applications where signal is limiting. These properties have also made this complex useful for single molecule and super-resolution microscopy.
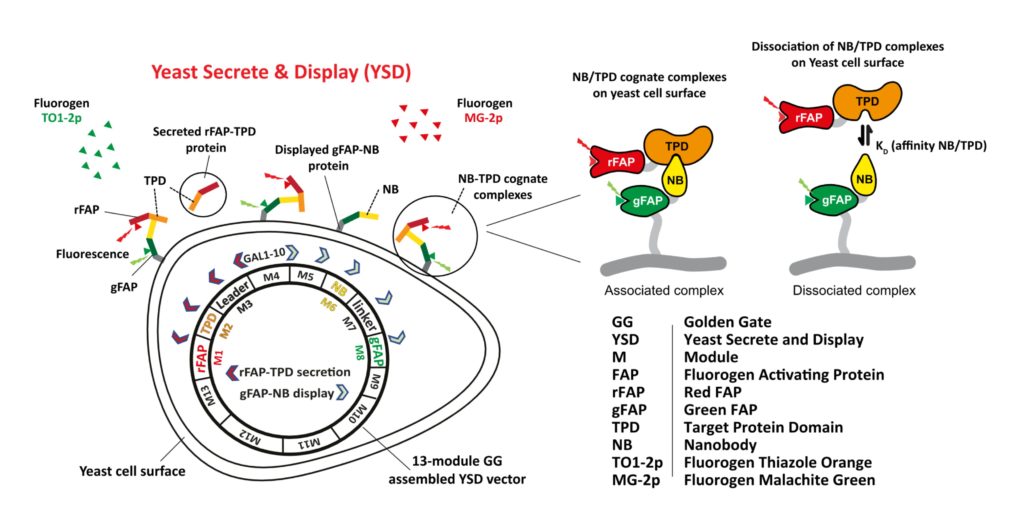
FAP-based labeling has been used for flow cytometric and plate-based small molecule drug discovery screens that monitor protein trafficking. As cartooned above and described under Mission and Technology, Biocognon is developing a Nanobody screening platform that employs two FAPs to monitor displayed Nanobodies and secreted target antigens expressed in a single yeast cell (ref. 152). The mechanism of the biosensor is not detailed here. We have demonstrated that our yeast system robustly co-expresses Nanobody-FAP and antigen-FAP fusions encoded by a modular vector that is optimized for synthetic biology. Nanobodies and target antigens that specifically associate on the cell surface can be flow cytometrically screened and identified using next generation sequencing analysis of barcoded output.
References
1. Szent-Gyorgyi C, Schmidt BF, Creeger Y, Fisher GW, Zakel KL, Adler S, et al. Fluorogen-activating single-chain antibodies for imaging cell surface proteins. Nat Biotechnol [Internet]. 2008 Feb;26(2):235–40. Available from: http://dx.doi.org/10.1038/nbt1368
2. Armitage BA, Berget PB. Chemistry. An enlightening structure-function relationship. Science [Internet]. 2008 Feb 29;319(5867):1195–6. Available from: http://dx.doi.org/10.1126/science.1155093
3. Evanko D. The new fluorescent probes on the block — Research Highlights. Nat Methods. 2008 Mar;5(3):218.
4. Ozhalici-Unal H, Pow CL, Marks SA, Jesper LD, Silva GL, Shank NI, et al. A rainbow of fluoromodules: a promiscuous scFv protein binds to and activates a diverse set of fluorogenic cyanine dyes. J Am Chem Soc [Internet]. 2008 Sep 24;130(38):12620–1. Available from: http://dx.doi.org/10.1021/ja805042p
5. Ediz V, Lee JL, Armitage BA, Yaron D. Molecular engineering of torsional potentials in fluorogenic dyes via electronic substituent effects. J Phys Chem A [Internet]. 2008 Oct 9;112(40):9692–701. Available from: http://dx.doi.org/10.1021/jp805546s
6. Bruchez M. State of the Art and Beyond: Fluorescent Probes for Living Cells. The Change We Need: New Frontiers in Live-Cell … [Internet]. 2009; Available from: https://www.researchgate.net/profile/Marcel_Bruchez/publication/255612997
7. Fitzpatrick JAJ, Yan Q, Sieber JJ, Dyba M, Schwarz U, Szent-Gyorgyi C, et al. STED nanoscopy in living cells using Fluorogen Activating Proteins. Bioconjug Chem [Internet]. 2009;20(10):1843–7. Available from: http://dx.doi.org/10.1021/bc900249e
8. Falco CN, Dykstra KM, Yates BP, Berget PB. scFv-based fluorogen activating proteins and variable domain inhibitors as fluorescent biosensor platforms. Biotechnol J [Internet]. 2009 Sep [cited 2022 Sep 9];4(9):1328–36. Available from: http://dx.doi.org/10.1002/biot.200900075
9. Shank NI, Zanotti KJ, Lanni F, Berget PB, Armitage BA. Enhanced photostability of genetically encodable fluoromodules based on fluorogenic cyanine dyes and a promiscuous protein partner. J Am Chem Soc [Internet]. 2009 Sep 16;131(36):12960–9. Available from: http://dx.doi.org/10.1021/ja9016864
10. Wu Y, Tapia PH, Gouveia K, Sklar LA, Fisher GW, Waggoner AS, et al. Inhibitors of FAP-fluorogen interaction as a multiplex assay tool compound for receptor internalization assays. In: Probe Reports from the NIH Molecular Libraries Program [Internet]. Bethesda (MD): National Center for Biotechnology Information (US); 2010 [cited 2024 Mar 25]. Available from: https://www.ncbi.nlm.nih.gov/pubmed/23833798
11. Fisher GW, Adler SA, Fuhrman MH, Waggoner AS, Bruchez MP, Jarvik JW. Detection and quantification of beta2AR internalization in living cells using FAP-based biosensor technology. J Biomol Screen [Internet]. 2010 Jul;15(6):703–9. Available from: http://dx.doi.org/10.1177/1087057110370892
12. Holleran J, Brown D, Fuhrman MH, Adler SA, Fisher GW, Jarvik JW. Fluorogen-activating proteins as biosensors of cell-surface proteins in living cells. Cytometry A [Internet]. 2010 Aug;77(8):776–82. Available from: http://dx.doi.org/10.1002/cyto.a.20925
13. Szent-Gyorgyi C, Schmidt BF, Fitzpatrick JAJ, Bruchez MP. Fluorogenic dendrons with multiple donor chromophores as bright genetically targeted and activated probes. J Am Chem Soc [Internet]. 2010 Aug 18;132(32):11103–9. Available from: http://dx.doi.org/10.1021/ja9099328
14. Sklar L, Edwards B. High throughput flow cytometry for discovery at UNMCMD and the NIH Molecular Libraries Initiative. Drug Discovery World. 2010 Oct 4;
15. Oprea TI, Bauman JE, Bologa CG, Buranda T, Chigaev A, Edwards BS, et al. Drug Repurposing from an Academic Perspective. Drug Discov Today Ther Strateg [Internet]. 2011;8(3–4):61–9. Available from: http://dx.doi.org/10.1016/j.ddstr.2011.10.002
16. Gaiotto T, Nguyen HB, Jung J, Gnanakaran GS, Schmidt JG, Waldo GS, et al. A photophysical study of two fluorogen-activating proteins bound to their cognate fluorogens. In: Enderlein J, Gryczynski ZK, Erdmann R, editors. Single Molecule Spectroscopy and Imaging IV [Internet]. SPIE; 2011. p. 79050O. (SPIE Proceedings; vol. 7905). Available from: http://proceedings.spiedigitallibrary.org/proceeding.aspx?doi=10.1117/12.875418
17. Zanotti KJ, Silva GL, Creeger Y, Robertson KL, Waggoner AS, Berget PB, et al. Blue fluorescent dye-protein complexes based on fluorogenic cyanine dyes and single chain antibody fragments. Org Biomol Chem [Internet]. 2011 Feb 21;9(4):1012–20. Available from: http://dx.doi.org/10.1039/c0ob00444h
18. Shruti S, Urban-Ciecko J, Fitzpatrick JA, Brenner R, Bruchez MP, Barth AL. The brain-specific Beta4 subunit downregulates BK channel cell surface expression. PLoS ONE [Internet]. 2012 Mar 16;7(3):e33429. Available from: http://dx.doi.org/10.1371/journal.pone.0033429
19. Senutovitch N, Stanfield RL, Bhattacharyya S, Rule GS, Wilson IA, Armitage BA, et al. A variable light domain fluorogen activating protein homodimerizes to activate dimethylindole red. Biochemistry [Internet]. 2012 Mar 27;51(12):2471–85. Available from: http://dx.doi.org/10.1021/bi201422g
20. Holleran JP, Glover ML, Peters KW, Bertrand CA, Watkins SC, Jarvik JW, et al. Pharmacological rescue of the mutant cystic fibrosis transmembrane conductance regulator (CFTR) detected by use of a novel fluorescence platform. Mol Med [Internet]. 2012 May 9;18:685–96. Available from: http://dx.doi.org/10.2119/molmed.2012.00001
21. Grover A, Schmidt BF, Salter RD, Watkins SC, Waggoner AS, Bruchez MP. Genetically encoded pH sensor for tracking surface proteins through endocytosis. Angew Chem Int Ed [Internet]. 2012 May 14 [cited 2021 Nov 6];51(20):4838–42. Available from: http://dx.doi.org/10.1002/anie.201108107
22. Müller T, Schumann C, Kraegeloh A. STED microscopy and its applications: new insights into cellular processes on the nanoscale. ChemPhysChem [Internet]. 2012 Jun 4;13(8):1986–2000. Available from: http://dx.doi.org/10.1002/cphc.201100986
23. Saunders MJ, Szent-Gyorgyi C, Fisher GW, Jarvik JW, Bruchez MP, Waggoner AS. Fluorogen activating proteins in flow cytometry for the study of surface molecules and receptors. Methods [Internet]. 2012 Jul;57(3):308–17. Available from: http://dx.doi.org/10.1016/j.ymeth.2012.02.003
24. Yushchenko DA, Zhang M, Yan Q, Waggoner AS, Bruchez MP. Genetically targetable and color-switching fluorescent probe. Chembiochem [Internet]. 2012 Jul 23;13(11):1564–8. Available from: http://dx.doi.org/10.1002/cbic.201200334
25. Kasuboski JM, Sigal YJ, Joens MS, Lillemeier BF, Fitzpatrick JAJ. Super‐resolution microscopy: A comparative treatment. Current Protocols in Cytometry [Internet]. 2012 Oct;62(1). Available from: https://currentprotocols.onlinelibrary.wiley.com/doi/10.1002/0471142956.cy0217s62
26. Wu Y, Tapia PH, Fisher GW, Simons PC, Strouse JJ, Foutz T, et al. Discovery of regulators of receptor internalization with high-throughput flow cytometry. Mol Pharmacol [Internet]. 2012 Oct;82(4):645–57. Available from: http://dx.doi.org/10.1124/mol.112.079897
27. Bachert C, Linstedt AD. A sensor of protein O-glycosylation based on sequential processing in the Golgi apparatus. Traffic [Internet]. 2013;14(1):47–56. Available from: http://dx.doi.org/10.1111/tra.12019
28. Shank NI, Pham HH, Waggoner AS, Armitage BA. Twisted cyanines: a non-planar fluorogenic dye with superior photostability and its use in a protein-based fluoromodule. J Am Chem Soc [Internet]. 2013 Jan 9 [cited 2021 Nov 3];135(1):242–51. Available from: http://dx.doi.org/10.1021/ja308629w
29. Wu Y, Tapia PH, Fisher GW, Waggoner AS, Jarvik J, Sklar LA. High-throughput flow cytometry compatible biosensor based on fluorogen activating protein technology. Cytometry A [Internet]. 2013 Feb;83(2):220–6. Available from: http://dx.doi.org/10.1002/cyto.a.22242
30. Saunders MJ, Liu W, Szent-Gyorgyi C, Wen Y, Drennen Z, Waggoner AS, et al. Engineering fluorogen activating proteins into self-assembling materials. Bioconjug Chem [Internet]. 2013 May 15 [cited 2021 Nov 4];24(5):803–10. Available from: http://dx.doi.org/10.1021/bc300613h
31. Holleran JP, Zeng J, Frizzell RA, Watkins SC. Regulated recycling of mutant CFTR is partially restored by pharmacological treatment. J Cell Sci [Internet]. 2013 Jun 15;126(Pt 12):2692–703. Available from: http://dx.doi.org/10.1242/jcs.120196
32. Yates BP, Peck MA, Berget PB. Directed evolution of a fluorogen-activating single chain antibody for function and enhanced brightness in the cytoplasm. Mol Biotechnol [Internet]. 2013 Jul;54(3):829–41. Available from: http://dx.doi.org/10.1007/s12033-012-9631-7
33. Xu J, Chang J, Yan Q, Dertinger T, Bruchez M, Weiss S. Labeling Cytosolic Targets in Live Cells with Blinking Probes. J Phys Chem Lett [Internet]. 2013 Jul 3;4(13):2138–46. Available from: http://dx.doi.org/10.1021/jz400682m
34. Szent-Gyorgyi C, Stanfield RL, Andreko S, Dempsey A, Ahmed M, Capek S, et al. Malachite green mediates homodimerization of antibody VL domains to form a fluorescent ternary complex with singular symmetric interfaces. J Mol Biol [Internet]. 2013 Nov 15;425(22):4595–613. Available from: http://dx.doi.org/10.1016/j.jmb.2013.08.014
35. Gallo E, Wienbar S, Snyder AC, Vasilev KV, Armitage BA, Jarvik JW. A single-chain-variable-fragment fluorescence biosensor activates fluorogens from dissimilar chemical families. Protein Pept Lett [Internet]. 2014;21(12):1289–94. Available from: http://dx.doi.org/10.2174/0929866521666140616121800
36. Saurabh S, Bruchez M. 15 targeting Dyes for Biology. Cell Membrane Nanodomains [Internet]. 2014; Available from: https://books.google.com/books?hl=en&lr=&id=BYLSBQAAQBAJ&oi=fnd&pg=PA341&dq=Marcel+P+Bruchez
37. Wu Y, Tapia PH, Jarvik J, Waggoner AS, Sklar LA. Real-time detection of protein trafficking with high-throughput flow cytometry (HTFC) and fluorogen-activating protein (FAP) base biosensor. Curr Protoc Cytom [Internet]. 2014 Jan 2;67:9.43.1-9.43.11. Available from: http://dx.doi.org/10.1002/0471142956.cy0943s67
38. Gallo E, Vasilev KV, Jarvik J. Fluorogen-activating-proteins as universal affinity biosensors for immunodetection. Biotechnol Bioeng [Internet]. 2014 Mar;111(3):475–84. Available from: http://dx.doi.org/10.1002/bit.25127
39. Yan Q, Schwartz SL, Maji S, Huang F, Szent-Gyorgyi C, Lidke DS, et al. Localization microscopy using noncovalent fluorogen activation by genetically encoded fluorogen-activating proteins. ChemPhysChem [Internet]. 2014 Mar 17;15(4):687–95. Available from: http://dx.doi.org/10.1002/cphc.201300757
40. Pastori C, Diomede L, Venuti A, Fisher G, Jarvik J, Bomsel M, et al. Induction of HIV-blocking anti-CCR5 IgA in Peyers’s patches without histopathological alterations. J Virol [Internet]. 2014 Apr;88(7):3623–35. Available from: http://dx.doi.org/10.1128/JVI.03663-13
41. Gallo E, Jarvik J. Fluorogen-activating scFv biosensors target surface markers on live cells via streptavidin or single-chain avidin. Mol Biotechnol [Internet]. 2014 Jul;56(7):585–90. Available from: http://dx.doi.org/10.1007/s12033-014-9732-6
42. Saunders MJ, Block E, Sorkin A, Waggoner AS, Bruchez MP. A bifunctional converter: fluorescein quenching scFv/fluorogen activating protein for photostability and improved signal to noise in fluorescence experiments. Bioconjug Chem [Internet]. 2014 Aug 20;25(8):1556–64. Available from: http://dx.doi.org/10.1021/bc500273n
43. Fisher GW, Fuhrman MH, Adler SA, Szent-Gyorgyi C, Waggoner AS, Jarvik JW. Self-Checking Cell-Based Assays for GPCR Desensitization and Resensitization. J Biomol Screen [Internet]. 2014 Sep;19(8):1220–6. Available from: http://dx.doi.org/10.1177/1087057114534299
44. Song L, Bachert C, Schjoldager KT, Clausen H, Linstedt AD. Development of isoform-specific sensors of polypeptide GalNAc-transferase activity. J Biol Chem [Internet]. 2014 Oct 31;289(44):30556–66. Available from: http://dx.doi.org/10.1074/jbc.M114.599563
45. Saurabh S, Beck LE, Maji S, Baty CJ, Wang Y, Yan Q, et al. Multiplexed modular genetic targeting of quantum dots. ACS Nano [Internet]. 2014 Nov 25;8(11):11138–46. Available from: http://dx.doi.org/10.1021/nn5044367
46. Yushchenko DA, Bruchez MP. Tailoring Fluorescent Labels for Far-Field Nanoscopy. In: Tinnefeld P, Eggeling C, Hell SW, editors. Far-Field Optical Nanoscopy [Internet]. Berlin, Heidelberg: Springer Berlin Heidelberg; 2015. p. 159–88. (Springer series on fluorescence: methods and applications; vol. 14). Available from: http://link.springer.com/10.1007/4243_2011_35
47. Wang Y, Telmer CA, Schmidt BF, Franke JD, Ort S, Arndt-Jovin DJ, et al. Fluorogen activating protein-affibody probes: modular, no-wash measurement of epidermal growth factor receptors. Bioconjug Chem [Internet]. 2015 Jan 21;26(1):137–44. Available from: http://dx.doi.org/10.1021/bc500525b
48. Schwartz SL, Yan Q, Telmer CA, Lidke KA, Bruchez MP, Lidke DS. Fluorogen-activating proteins provide tunable labeling densities for tracking FcεRI independent of IgE. ACS Chem Biol [Internet]. 2015 Feb 20;10(2):539–46. Available from: http://dx.doi.org/10.1021/cb5005146
49. Yan Q, Schmidt BF, Perkins LA, Naganbabu M, Saurabh S, Andreko SK, et al. Near-instant surface-selective fluorogenic protein quantification using sulfonated triarylmethane dyes and fluorogen activating proteins. Org Biomol Chem [Internet]. 2015 Feb 21;13(7):2078–86. Available from: http://dx.doi.org/10.1039/c4ob02309a
50. Pham HH, Szent-Gyorgyi C, Brotherton WL, Schmidt BF, Zanotti KJ, Waggoner AS, et al. Bichromophoric dyes for wavelength shifting of dye-protein fluoromodules. Org Biomol Chem [Internet]. 2015 Mar 28;13(12):3699–710. Available from: http://dx.doi.org/10.1039/c4ob02522a
51. Yan Q, Bruchez MP. Advances in chemical labeling of proteins in living cells. Cell Tissue Res [Internet]. 2015 Apr [cited 2021 Nov 6];360(1):179–94. Available from: http://dx.doi.org/10.1007/s00441-015-2145-4
52. Telmer CA, Verma R, Teng H, Andreko S, Law L, Bruchez MP. Rapid, specific, no-wash, far-red fluorogen activation in subcellular compartments by targeted fluorogen activating proteins. ACS Chem Biol [Internet]. 2015 May 15;10(5):1239–46. Available from: http://dx.doi.org/10.1021/cb500957k
53. Senutovitch N, Vernetti L, Boltz R, DeBiasio R, Gough A, Taylor DL. Fluorescent protein biosensors applied to microphysiological systems. Exp Biol Med (Maywood) [Internet]. 2015 Jun [cited 2024 Mar 25];240(6):795–808. Available from: http://dx.doi.org/10.1177/1535370215584934
54. Edwards BS, Sklar LA. Flow cytometry: impact on early drug discovery. J Biomol Screen [Internet]. 2015 Jul;20(6):689–707. Available from: http://dx.doi.org/10.1177/1087057115578273
55. Bruchez MP. Dark dyes-bright complexes: fluorogenic protein labeling. Curr Opin Chem Biol [Internet]. 2015 Aug [cited 2021 Nov 6];27:18–23. Available from: http://dx.doi.org/10.1016/j.cbpa.2015.05.014
56. Pratt CP, He J, Wang Y, Barth AL, Bruchez MP. Fluorogenic Green-Inside Red-Outside (GIRO) Labeling Approach Reveals Adenylyl Cyclase-Dependent Control of BKα Surface Expression. Bioconjug Chem [Internet]. 2015 Sep 16;26(9):1963–71. Available from: http://dx.doi.org/10.1021/acs.bioconjchem.5b00409
57. Rastede EE, Tanha M, Yaron D, Watkins SC, Waggoner AS, Armitage BA. Spectral fine tuning of cyanine dyes: electron donor-acceptor substituted analogues of thiazole orange. Photochem Photobiol Sci [Internet]. 2015 Sep 26;14(9):1703–12. Available from: http://dx.doi.org/10.1039/c5pp00117j
58. Gallo E, Snyder AC, Jarvik JW. Engineering tandem single-chain Fv as cell surface reporters with enhanced properties of fluorescence detection. Protein Eng Des Sel [Internet]. 2015 Oct [cited 2021 Nov 6];28(10):327–37. Available from: http://dx.doi.org/10.1093/protein/gzv016
59. Magenau AJD, Saurabh S, Andreko SK, Telmer CA, Schmidt BF, Waggoner AS, et al. Genetically targeted fluorogenic macromolecules for subcellular imaging and cellular perturbation. Biomaterials [Internet]. 2015 Oct;66:1–8. Available from: http://dx.doi.org/10.1016/j.biomaterials.2015.07.002
60. Zhang M, Chakraborty SK, Sampath P, Rojas JJ, Hou W, Saurabh S, et al. Fluoromodule-based reporter/probes designed for in vivo fluorescence imaging. J Clin Invest [Internet]. 2015 Oct;125(10):3915–27. Available from: http://dx.doi.org/10.1172/JCI81086
61. Saurabh S, Zhang M, Mann VR, Costello AM, Bruchez MP. Kinetically Tunable Photostability of Fluorogen-Activating Peptide-Fluorogen Complexes. ChemPhysChem [Internet]. 2015 Oct 5;16(14):2974–80. Available from: http://dx.doi.org/10.1002/cphc.201500587
62. Snyder JC, Pack TF, Rochelle LK, Chakraborty SK, Zhang M, Eaton AW, et al. A rapid and affordable screening platform for membrane protein trafficking. BMC Biol [Internet]. 2015 Dec 17;13:107. Available from: http://dx.doi.org/10.1186/s12915-015-0216-3
63. Song L, Bachert C, Linstedt AD. Activity Detection of GalNAc Transferases by Protein-Based Fluorescence Sensors In Vivo. Methods Mol Biol [Internet]. 2016 [cited 2022 Sep 9];1496:123–31. Available from: http://dx.doi.org/10.1007/978-1-4939-6463-5_10
64. Vasilev KV, Gallo E, Shank N, Jarvik JW. Novel biosensor of membrane protein proximity based on fluorogen activated proteins. Comb Chem High Throughput Screen [Internet]. 2016;19(5):392–9. Available from: http://dx.doi.org/10.2174/1386207319666160408150320
65. Wu Y, Stauffer SR, Stanfield RL, Tapia PH, Ursu O, Fisher GW, et al. Discovery of Small-Molecule Nonfluorescent Inhibitors of Fluorogen-Fluorogen Activating Protein Binding Pair. J Biomol Screen [Internet]. 2016;21(1):74–87. Available from: http://dx.doi.org/10.1177/1087057115609145
66. He J, Wang Y, Missinato MA, Onuoha E, Perkins LA, Watkins SC, et al. A genetically targetable near-infrared photosensitizer. Nat Methods [Internet]. 2016 Mar;13(3):263–8. Available from: http://dx.doi.org/10.1038/nmeth.3735
67. Larsen MB, Hu J, Frizzell RA, Watkins SC. Simple image-based no-wash method for quantitative detection of surface expressed CFTR. Methods [Internet]. 2016 Mar;96:40–5. Available from: http://dx.doi.org/10.1016/j.ymeth.2015.09.006
68. Armitage BA. Fluoromodules: Fluorescent Dye-Protein Complexes for Genetically Encodable Labels. In: Wilhelmsson M, Tor Y, editors. Fluorescent analogs of biomolecular building blocks: design and applications [Internet]. Hoboken, NJ, USA: John Wiley & Sons, Inc; 2016. p. 124–36. Available from: https://onlinelibrary.wiley.com/doi/10.1002/9781119179320.ch6
69. Schütz M, Batyuk A, Klenk C, Kummer L, de Picciotto S, Gülbakan B, et al. Generation of Fluorogen-Activating Designed Ankyrin Repeat Proteins (FADAs) as Versatile Sensor Tools. J Mol Biol [Internet]. 2016 Mar 27;428(6):1272–89. Available from: http://dx.doi.org/10.1016/j.jmb.2016.01.017
70. Backlund MP, Arbabi A, Petrov PN, Arbabi E, Saurabh S, Faraon A, et al. Removing Orientation-Induced Localization Biases in Single-Molecule Microscopy Using a Broadband Metasurface Mask. Nat Photonics [Internet]. 2016 May 16;10:459–62. Available from: http://dx.doi.org/10.1038/nphoton.2016.93
71. Liu W, Saunders MJ, Bagia C, Freeman EC, Fan Y, Gawalt ES, et al. Local retention of antibodies in vivo with an injectable film embedded with a fluorogen-activating protein. J Control Release [Internet]. 2016 May 28;230:1–12. Available from: http://dx.doi.org/10.1016/j.jconrel.2016.03.032
72. Naganbabu M, Perkins LA, Wang Y, Kurish J, Schmidt BF, Bruchez MP. Multiexcitation fluorogenic labeling of surface, intracellular, and total protein pools in living cells. Bioconjug Chem [Internet]. 2016 Jun 15;27(6):1525–31. Available from: http://dx.doi.org/10.1021/acs.bioconjchem.6b00169
73. Kagan VE, Jiang J, Huang Z, Tyurina YY, Desbourdes C, Cottet-Rousselle C, et al. NDPK-D (NM23-H4)-mediated externalization of cardiolipin enables elimination of depolarized mitochondria by mitophagy. Cell Death Differ [Internet]. 2016 Jul [cited 2024 Mar 25];23(7):1140–51. Available from: http://dx.doi.org/10.1038/cdd.2015.160
74. Saurabh S, Perez AM, Comerci CJ, Shapiro L, Moerner WE. Super-resolution Imaging of Live Bacteria Cells Using a Genetically Directed, Highly Photostable Fluoromodule. J Am Chem Soc [Internet]. 2016 Aug 24;138(33):10398–401. Available from: http://dx.doi.org/10.1021/jacs.6b05943
75. Helfield B, Chen X, Watkins SC, Villanueva FS. Biophysical insight into mechanisms of sonoporation. Proc Natl Acad Sci USA [Internet]. 2016 Sep 6 [cited 2024 Mar 25];113(36):9983–8. Available from: http://dx.doi.org/10.1073/pnas.1606915113
76. Kagan VE, Mao G, Qu F, Angeli JPF, Doll S, Croix CS, et al. Oxidized arachidonic and adrenic PEs navigate cells to ferroptosis. Nat Chem Biol [Internet]. 2017;13(1):81–90. Available from: http://dx.doi.org/10.1038/nchembio.2238
77. Wang Y, Ballou B, Schmidt BF, Andreko S, St Croix CM, Watkins SC, et al. Affibody-targeted fluorogen activating protein for in vivo tumor imaging. Chem Commun [Internet]. 2017 Feb 7 [cited 2021 Nov 6];53(12):2001–4. Available from: http://dx.doi.org/10.1039/c6cc09137g
78. Boeck JM, Spencer JV. Effect of human cytomegalovirus (HCMV) US27 on CXCR4 receptor internalization measured by fluorogen-activating protein (FAP) biosensors. PLoS ONE [Internet]. 2017 Feb 16;12(2):e0172042. Available from: http://dx.doi.org/10.1371/journal.pone.0172042
79. Ho HC, MacGurn JA, Emr SD. Deubiquitinating enzymes Ubp2 and Ubp15 regulate endocytosis by limiting ubiquitination and degradation of ARTs. Mol Biol Cell [Internet]. 2017 Mar 15;28(9):1271–83. Available from: http://dx.doi.org/10.1091/mbc.E17-01-0008
80. Song L, Linstedt AD. Inhibitor of ppGalNAc-T3-mediated O-glycosylation blocks cancer cell invasiveness and lowers FGF23 levels. eLife [Internet]. 2017 Mar 31;6. Available from: http://dx.doi.org/10.7554/eLife.24051
81. Thorn K. Genetically encoded fluorescent tags. Mol Biol Cell [Internet]. 2017 Apr;28(7):848–57. Available from: http://dx.doi.org/10.1091/mbc.E16-07-0504
82. Snyder JC, Rochelle LK, Ray C, Pack TF, Bock CB, Lubkov V, et al. Inhibiting clathrin-mediated endocytosis of the leucine-rich G protein-coupled receptor-5 diminishes cell fitness. J Biol Chem [Internet]. 2017 Apr 28;292(17):7208–22. Available from: http://dx.doi.org/10.1074/jbc.M116.756635
83. Xie M, Tang S, Li K, Ding S. Pharmacological reprogramming of somatic cells for regenerative medicine. Acc Chem Res [Internet]. 2017 May 16 [cited 2024 Mar 25];50(5):1202–11. Available from: http://dx.doi.org/10.1021/acs.accounts.7b00020
84. Ackerman DS, Vasilev KV, Schmidt BF, Cohen LB, Jarvik JW. Tethered fluorogen assay to visualize membrane apposition in living cells. Bioconjug Chem [Internet]. 2017 May 17;28(5):1356–62. Available from: http://dx.doi.org/10.1021/acs.bioconjchem.7b00047
85. Saurabh S, Perez AM, Comerci CJ, Shapiro L, Moerner WE. Super-Resolution Microscopy and Single-Protein Tracking in Live Bacteria Using a Genetically Encoded, Photostable Fluoromodule. Curr Protoc Cell Biol [Internet]. 2017 Jun 19;75:4.32.1-4.32.22. Available from: http://dx.doi.org/10.1002/cpcb.21
86. Pena K, Larsen M, Calderon M, Tsang M, Watkins SC, Bruchez MP, et al. Combining novel probes and high resolution imaging to dissect mitochondrial function in living systems. Microsc Microanal [Internet]. 2017 Jul;23(S1):1170–1. Available from: https://www.cambridge.org/core/product/identifier/S1431927617006511/type/journal_article
87. Li C, Tebo AG, Gautier A. Fluorogenic labeling strategies for biological imaging. Int J Mol Sci [Internet]. 2017 Jul 9;18(7). Available from: http://dx.doi.org/10.3390/ijms18071473
88. Gallo E, Jarvik J. Breaking the color barrier – a multi-selective antibody reporter offers innovative strategies of fluorescence detection. J Cell Sci [Internet]. 2017 Aug [cited 2022 Sep 12];130(15):2644–53. Available from: http://dx.doi.org/10.1242/jcs.202952
89. Shiwarski DJ, Darr M, Telmer CA, Bruchez MP, Puthenveedu MA. PI3K class II α regulates δ-opioid receptor export from the trans-Golgi network. Mol Biol Cell [Internet]. 2017 Aug;28(16):2202–19. Available from: http://dx.doi.org/10.1091/mbc.E17-01-0030
90. Zhang Q, Wang Q, Sun Y, Zuo L, Fetz V, Hu HY. Superior Fluorogen-Activating Protein Probes Based on 3-Indole-Malachite Green. Org Lett [Internet]. 2017 Sep;19(17):4496–9. Available from: http://dx.doi.org/10.1021/acs.orglett.7b02055
91. Pratt CP, Kuljis DA, Homanics GE, He J, Kolodieznyi D, Dudem S, et al. Tagging of Endogenous BK Channels with a Fluorogen-Activating Peptide Reveals β4-Mediated Control of Channel Clustering in Cerebellum. Front Cell Neurosci [Internet]. 2017 Oct 31 [cited 2021 Nov 6];11:337. Available from: http://dx.doi.org/10.3389/fncel.2017.00337
92. Lorenz-Guertin JM, Wilcox MR, Zhang M, Larsen MB, Pilli J, Schmidt BF, et al. A versatile optical tool for studying synaptic GABAA receptor trafficking. J Cell Sci [Internet]. 2017 Nov 15;130(22):3933–45. Available from: http://dx.doi.org/10.1242/jcs.205286
93. Ferreira K. Synthesis of siderophore-based conjugates to detect and treat bacterial infections. 2018; Available from: https://www.repo.uni-hannover.de/handle/123456789/3603
94. Giuliano KA, Wachi S, Drew L, Dukovski D, Green O, Bastos C, et al. Use of a High-Throughput Phenotypic Screening Strategy to Identify Amplifiers, a Novel Pharmacological Class of Small Molecules That Exhibit Functional Synergy with Potentiators and Correctors. SLAS Discov [Internet]. 2018 Feb;23(2):111–21. Available from: http://dx.doi.org/10.1177/2472555217729790
95. Perkins LA, Yan Q, Schmidt BF, Kolodieznyi D, Saurabh S, Larsen MB, et al. Genetically targeted ratiometric and activated ph indicator complexes (traphic) for receptor trafficking. Biochemistry [Internet]. 2018 Feb 6;57(5):861–71. Available from: http://dx.doi.org/10.1021/acs.biochem.7b01135
96. Kirschman J, Qi M, Ding L, Hammonds J, Dienger-Stambaugh K, Wang JJ, et al. HIV-1 Envelope Glycoprotein Trafficking through the Endosomal Recycling Compartment Is Required for Particle Incorporation. J Virol [Internet]. 2018 Mar;92(5). Available from: http://dx.doi.org/10.1128/JVI.01893-17
97. Meier JL, Schnermann MJ. Pharmacology by chemical biology. Mol Pharm [Internet]. 2018 Mar 5;15(3):703–4. Available from: http://dx.doi.org/10.1021/acs.molpharmaceut.8b00067
98. Perkins LA, Fisher GW, Naganbabu M, Schmidt BF, Mun F, Bruchez MP. High-Content Surface and Total Expression siRNA Kinase Library Screen with VX-809 Treatment Reveals Kinase Targets that Enhance F508del-CFTR Rescue. Mol Pharm [Internet]. 2018 Mar 5;15(3):759–67. Available from: http://dx.doi.org/10.1021/acs.molpharmaceut.7b00928
99. Braun A, Farber MJ, Klase ZA, Berget PB, Myers KA. A cell surface display fluorescent biosensor for measuring MMP14 activity in real-time. Sci Rep [Internet]. 2018 Apr 12;8(1):5916. Available from: http://dx.doi.org/10.1038/s41598-018-24080-0
100. Xu S, Hu HY. Fluorogen-activating proteins: beyond classical fluorescent proteins. Acta Pharm Sin B [Internet]. 2018 May;8(3):339–48. Available from: http://linkinghub.elsevier.com/retrieve/pii/S221138351730597X
101. Hager NA, Krasowski CJ, Mackie TD, Kolb AR, Needham PG, Augustine AA, et al. Select α-arrestins control cell-surface abundance of the mammalian Kir2.1 potassium channel in a yeast model. J Biol Chem [Internet]. 2018 Jul 13;293(28):11006–21. Available from: http://dx.doi.org/10.1074/jbc.RA117.001293
102. Ding M, Clark R, Bardelle C, Backmark A, Norris T, Williams W, et al. Application of High-Throughput Flow Cytometry in Early Drug Discovery: An AstraZeneca Perspective. SLAS Discov [Internet]. 2018 Aug;23(7):719–31. Available from: http://dx.doi.org/10.1177/2472555218775074
103. Mackie DI, Al Mutairi F, Davis RB, Kechele DO, Nielsen NR, Snyder JC, et al. hCALCRL mutation causes autosomal recessive nonimmune hydrops fetalis with lymphatic dysplasia. J Exp Med [Internet]. 2018 Sep 3;215(9):2339–53. Available from: http://dx.doi.org/10.1084/jem.20180528
104. Zeng G, Wang Y, Bruchez MP, Liang FS. Self-Reporting Chemically Induced Protein Proximity System Based on a Malachite Green Derivative and the L5** Fluorogen Activating Protein. Bioconjug Chem [Internet]. 2018 Sep 19;29(9):3010–5. Available from: http://dx.doi.org/10.1021/acs.bioconjchem.8b00415
105. Emmerstorfer-Augustin A, Augustin CM, Shams S, Thorner J. Tracking yeast pheromone receptor Ste2 endocytosis using fluorogen-activating protein tagging. Mol Biol Cell [Internet]. 2018 Nov;29(22):2720–36. Available from: http://dx.doi.org/10.1091/mbc.E18-07-0424
106. Bozhanova NG, Baranov MS, Baleeva NS, Gavrikov AS, Mishin AS. Red-Shifted Aminated Derivatives of GFP Chromophore for Live-Cell Protein Labeling with Lipocalins. Int J Mol Sci [Internet]. 2018 Nov 28;19(12). Available from: http://dx.doi.org/10.3390/ijms19123778
107. Martynov VI, Pakhomov AA, Deyev IE, Petrenko AG. Genetically encoded fluorescent indicators for live cell pH imaging. Biochim Biophys Acta Gen Subj [Internet]. 2018 Dec;1862(12):2924–39. Available from: http://dx.doi.org/10.1016/j.bbagen.2018.09.013
108. Hintze J, Ye Z, Narimatsu Y, Madsen TD, Joshi HJ, Goth CK, et al. Probing the contribution of individual polypeptide GalNAc-transferase isoforms to the O-glycoproteome by inducible expression in isogenic cell lines. J Biol Chem [Internet]. 2018 Dec 7 [cited 2022 Sep 9];293(49):19064–77. Available from: http://dx.doi.org/10.1074/jbc.RA118.004516
109. Chen W, Shen B, Sun X. Analysis of Progress and Challenges of EGFR-Targeted Molecular Imaging in Cancer With a Focus on Affibody Molecules. Mol Imaging [Internet]. 2019 [cited 2024 Mar 25];18:1536012118823473. Available from: http://dx.doi.org/10.1177/1536012118823473
110. Gong X, Liao Y, Ahner A, Larsen MB, Wang X, Bertrand CA, et al. Different SUMO paralogues determine the fate of wild-type and mutant CFTRs: biogenesis versus degradation. Mol Biol Cell [Internet]. 2019;30(1):4–16. Available from: http://dx.doi.org/10.1091/mbc.E18-04-0252
111. Mishin AS, Lukyanov KA. Live-Cell Super-resolution Fluorescence Microscopy. Biochemistry Mosc [Internet]. 2019;84(Suppl 1):S19–31. Available from: http://dx.doi.org/10.1134/S0006297919140025
112. Perez Verdaguer M, Larsen M, Bruchez M, Watkins S, Sorkin A. Analysis of EGF receptor endocytosis using fluorogen activating protein tagged receptor. Bio Protoc [Internet]. 2019 [cited 2021 Nov 6];9(24). Available from: https://bio-protocol.org/e3463
113. Stensrud KF, Zanotti KJ, Waggoner AS, Armitage BA. Spectral Properties of Fluorogenic Thiophene-derived Triarylmethane Dyes. Photochem Photobiol [Internet]. 2019;95(1):406–10. Available from: http://dx.doi.org/10.1111/php.13037
114. Kozma E, Kele P. Fluorogenic probes for super-resolution microscopy. Org Biomol Chem [Internet]. 2019 Jan 2;17(2):215–33. Available from: http://dx.doi.org/10.1039/c8ob02711k
115. Ackerman DS, Altun B, Kolodieznyi D, Bruchez MP, Tsourkas A, Jarvik JW. Antibody-Linked Fluorogen-Activating Proteins for Antigen Detection and Cell Ablation. Bioconjug Chem [Internet]. 2019 Jan 16;30(1):63–9. Available from: http://dx.doi.org/10.1021/acs.bioconjchem.8b00720
116. Manoukian R, Sun H, Miller S, Shi D, Chan B, Xu C. Effects of monoclonal antagonist antibodies on calcitonin gene-related peptide receptor function and trafficking. J Headache Pain [Internet]. 2019 Apr 30;20(1):44. Available from: http://dx.doi.org/10.1186/s10194-019-0992-1
117. Larsen MB, Perez Verdaguer M, Schmidt BF, Bruchez MP, Watkins SC, Sorkin A. Generation of endogenous pH-sensitive EGF receptor and its application in high-throughput screening for proteins involved in clathrin-mediated endocytosis. eLife [Internet]. 2019 May 8;8. Available from: https://elifesciences.org/articles/46135
118. Ayele TM, Knutson SD, Ellipilli S, Hwang H, Heemstra JM. Fluorogenic photoaffinity labeling of proteins in living cells. Bioconjug Chem [Internet]. 2019 May 15 [cited 2024 Mar 25];30(5):1309–13. Available from: http://dx.doi.org/10.1021/acs.bioconjchem.9b00203
119. Xiong Y, Tian X, Ai HW. Molecular tools to generate reactive oxygen species in biological systems. Bioconjug Chem [Internet]. 2019 May 15;30(5):1297–303. Available from: http://dx.doi.org/10.1021/acs.bioconjchem.9b00191
120. Lam WY, Wang Y, Mostofian B, Jorgens D, Kwon S, Chin K, et al. HER2 cancer protrusion growth signaling regulated by unhindered, localized filopodial dynamics. BioRxiv [Internet]. 2019 Jun 2 [cited 2024 Mar 25]; Available from: http://biorxiv.org/lookup/doi/10.1101/654988
121. Fouquerel E, Barnes RP, Uttam S, Watkins SC, Bruchez MP, Opresko PL. Targeted and Persistent 8-Oxoguanine Base Damage at Telomeres Promotes Telomere Loss and Crisis. Mol Cell [Internet]. 2019 Jul 11;75(1):117-130.e6. Available from: http://dx.doi.org/10.1016/j.molcel.2019.04.024
122. Jang S, Kumar N, Beckwitt EC, Kong M, Fouquerel E, Rapić-Otrin V, et al. Damage sensor role of UV-DDB during base excision repair. Nat Struct Mol Biol [Internet]. 2019 Aug;26(8):695–703. Available from: http://www.nature.com/articles/s41594-019-0261-7
123. Bulgari D, Deitcher DL, Schmidt BF, Carpenter MA, Szent-Gyorgyi C, Bruchez MP, et al. Activity-evoked and spontaneous opening of synaptic fusion pores. Proc Natl Acad Sci USA [Internet]. 2019 Aug 20;116(34):17039–44. Available from: http://dx.doi.org/10.1073/pnas.1905322116
124. Qian W, Kumar N, Roginskaya V, Fouquerel E, Opresko PL, Shiva S, et al. Chemoptogenetic damage to mitochondria causes rapid telomere dysfunction. Proc Natl Acad Sci USA [Internet]. 2019 Sep 10;116(37):18435–44. Available from: http://dx.doi.org/10.1073/pnas.1910574116
125. Whinn KS, Kaur G, Lewis JS, Schauer GD, Mueller SH, Jergic S, et al. Nuclease dead Cas9 is a programmable roadblock for DNA replication. Sci Rep [Internet]. 2019 Sep 16;9(1):13292. Available from: http://www.nature.com/articles/s41598-019-49837-z
126. Litvina E, Adams A, Barth A, Bruchez M, Carson J, Chung JE, et al. BRAIN Initiative: Cutting-Edge Tools and Resources for the Community. J Neurosci [Internet]. 2019 Oct 16;39(42):8275–84. Available from: http://www.jneurosci.org/lookup/doi/10.1523/JNEUROSCI.1169-19.2019
127. Kuljis DA, Park E, Telmer CA, Lee J, Ackerman DS, Bruchez MP, et al. Fluorescence-Based Quantitative Synapse Analysis for Cell Type-Specific Connectomics. eNeuro [Internet]. 2019 Oct 31;6(5). Available from: http://dx.doi.org/10.1523/ENEURO.0193-19.2019
128. Gallo E. Fluorogen-Activating Proteins: Next-Generation Fluorescence Probes for Biological Research. Bioconjug Chem [Internet]. 2020 Jan 15;31(1):16–27. Available from: http://dx.doi.org/10.1021/acs.bioconjchem.9b00710
129. Xie W, Jiao B, Bai Q, Ilin VA, Sun M, Burton CE, et al. Chemoptogenetic ablation of neuronal mitochondria in vivo with spatiotemporal precision and controllable severity. eLife [Internet]. 2020 Mar 17;9. Available from: https://elifesciences.org/articles/51845
130. Perkins LA, Bruchez MP. Fluorogen activating protein toolset for protein trafficking measurements. Traffic [Internet]. 2020 Apr;21(4):333–48. Available from: http://dx.doi.org/10.1111/tra.12722
131. Ayele TM, Knutson SD, Heemstra JM. Covalent live-cell labeling of proteins using a photoreactive fluorogen. Meth Enzymol [Internet]. 2020 Apr 28;639:355–77. Available from: http://dx.doi.org/10.1016/bs.mie.2020.04.019
132. Helfield B, Chen X, Watkins SC, Villanueva FS. Transendothelial Perforations and the Sphere of Influence of Single-Site Sonoporation. Ultrasound Med Biol [Internet]. 2020 Jul;46(7):1686–97. Available from: http://dx.doi.org/10.1016/j.ultrasmedbio.2020.02.017
133. Carpenter MA, Wang Y, Telmer CA, Schmidt BF, Yang Z, Bruchez MP. Protein Proximity Observed Using Fluorogen Activating Protein and Dye Activated by Proximal Anchoring (FAP-DAPA) System. ACS Chem Biol [Internet]. 2020 Sep 18;15(9):2433–43. Available from: https://pubs.acs.org/doi/10.1021/acschembio.0c00419
134. Liang P, Kolodieznyi D, Creeger Y, Ballou B, Bruchez MP. Subcellular singlet oxygen and cell death: location matters. Front Chem [Internet]. 2020 Nov 17 [cited 2021 Nov 6];8:592941. Available from: https://www.frontiersin.org/articles/10.3389/fchem.2020.592941/full
135. Gu B, Comerci CJ, McCarthy DG, Saurabh S, Moerner WE, Wysocka J. Opposing Effects of Cohesin and Transcription on CTCF Organization Revealed by Super-resolution Imaging. Mol Cell [Internet]. 2020 Nov 19;80(4):699-711.e7. Available from: https://linkinghub.elsevier.com/retrieve/pii/S1097276520306845
136. Dichmann L, Bregnhøj M, Liu H, Westberg M, Poulsen TB, Etzerodt M, et al. Photophysics of a protein-bound derivative of malachite green that sensitizes the production of singlet oxygen. Photochem Photobiol Sci [Internet]. 2021 Mar 15;20(3):435–49. Available from: http://dx.doi.org/10.1007/s43630-021-00032-y
137. Zhong Z, Ning J, Boggs EA, Jang S, Wallace C, Telmer C, et al. Cytoplasmic CPSF6 Regulates HIV-1 Capsid Trafficking and Infection in a Cyclophilin A-Dependent Manner. MBio [Internet]. 2021 Mar 23;12(2). Available from: http://dx.doi.org/10.1128/mBio.03142-20
138. Klose MK, Bruchez MP, Deitcher DL, Levitan ES. Temporally and spatially partitioned neuropeptide release from individual clock neurons. Proc Natl Acad Sci USA [Internet]. 2021 Apr 27;118(17). Available from: http://dx.doi.org/10.1073/pnas.2101818118
139. Kataoka S, Manandhar P, Lee J, Workman CJ, Banerjee H, Szymczak-Workman AL, et al. The costimulatory activity of Tim-3 requires Akt and MAPK signaling and its recruitment to the immune synapse. Sci Signal [Internet]. 2021 Jun 15;14(687). Available from: https://stke.sciencemag.org/lookup/doi/10.1126/scisignal.aba0717
140. Missinato MA, Zuppo DA, Watkins SC, Bruchez MP, Tsang M. Zebrafish heart regenerates after chemoptogenetic cardiomyocyte depletion. Dev Dyn [Internet]. 2021 Jul;250(7):986–1000. Available from: http://dx.doi.org/10.1002/dvdy.305
141. Perez Verdaguer M, Zhang T, Paulo JA, Gygi S, Watkins SC, Sakurai H, et al. Mechanism of p38 MAPK-induced EGFR endocytosis and its crosstalk with ligand-induced pathways. J Cell Biol [Internet]. 2021 Jul 5;220(7). Available from: http://dx.doi.org/10.1083/jcb.202102005
142. Goeckeler-Fried JL, Aldrin Denny R, Joshi D, Hill C, Larsen MB, Chiang AN, et al. Improved correction of F508del-CFTR biogenesis with a folding facilitator and an inhibitor of protein ubiquitination. Bioorg Med Chem Lett [Internet]. 2021 Sep 15;48:128243. Available from: https://linkinghub.elsevier.com/retrieve/pii/S0960894X21004704
143. Wang X, Wang Q, Zhang Q, Han X, Xu S, Yin D, et al. Developing fluoromodule-based probes for in vivo monitoring the bacterial infections and antibiotic responses. Talanta [Internet]. 2021 Oct;233:122610. Available from: http://dx.doi.org/10.1016/j.talanta.2021.122610
144. Larsen MB, Choi JJ, Wang X, Myerburg MM, Frizzell RA, Bertrand CA. Separating the contributions of SLC26A9 and CFTR to anion secretion in primary human bronchial epithelia. Am J Physiol Lung Cell Mol Physiol [Internet]. 2021 Dec;321(6):L1147–60. Available from: http://dx.doi.org/10.1152/ajplung.00563.2020
145. Wang L, Lu Z, Zhao J, Schank M, Cao D, Dang X, et al. Selective oxidative stress induces dual damage to telomeres and mitochondria in human T cells. Aging Cell [Internet]. 2021 Dec;20(12):e13513. Available from: http://dx.doi.org/10.1111/acel.13513
146. Saurabh S, Chong TN, Bayas C, Dahlberg PD, Cartwright HN, Moerner WE, et al. ATP-responsive biomolecular condensates tune bacterial kinase signaling. Sci Adv [Internet]. 2022 Feb 18;8(7):eabm6570. Available from: http://dx.doi.org/10.1126/sciadv.abm6570
147. He J, Zhou Y, Liu Y, Guo R, Jiang J, Bruchez MP. Fluorogen-Activating-Protein-Loaded Tantalum Oxide Nanoshells for in Vivo On-Demand Fluorescence/Photoacoustic Imaging. ACS Appl Bio Mater [Internet]. 2022 Mar 21;5(3):1057–63. Available from: http://dx.doi.org/10.1021/acsabm.1c01113
148. Guy C, Mitrea DM, Chou PC, Temirov J, Vignali KM, Liu X, et al. LAG3 associates with TCR-CD3 complexes and suppresses signaling by driving co-receptor-Lck dissociation. Nat Immunol [Internet]. 2022 May;23(5):757–67. Available from: http://dx.doi.org/10.1038/s41590-022-01176-4
149. Barnes RP, de Rosa M, Thosar SA, Detwiler AC, Roginskaya V, Van Houten B, et al. Telomeric 8-oxo-guanine drives rapid premature senescence in the absence of telomere shortening. Nat Struct Mol Biol [Internet]. 2022 Jul;29(7):639–52. Available from: https://www.nature.com/articles/s41594-022-00790-y
150. Romesberg A, Van Houten B. Targeting Mitochondrial Function with Chemoptogenetics. Biomedicines [Internet]. 2022 Oct;10(10). Available from: http://dx.doi.org/10.3390/biomedicines10102459
151. Guo Y, Song M, Liu X, Chen Y, Xun Z, Sun Y, et al. Photodynamic therapy-improved oncolytic bacterial immunotherapy with FAP-encoding S. typhimurium. J Control Release [Internet]. 2022 Nov;351:860–71. Available from: http://dx.doi.org/10.1016/j.jconrel.2022.09.050
152. Szent-Gyorgyi C, Perkins LA, Schmidt BF, Liu Z, Bruchez MP, van de Weerd R. Bottom-Up Design: A Modular Golden Gate Assembly Platform of Yeast Plasmids for Simultaneous Secretion and Surface Display of Distinct FAP Fusion Proteins. ACS Synth Biol [Internet]. 2022 Nov 18;11(11):3681–98. Available from: http://dx.doi.org/10.1021/acssynbio.2c00283
153. Han S, Sims A, Aceto A, Schmidt BF, Bruchez MP, Gurkar AU. A chemoptogenetic tool for spatiotemporal induction of oxidative DNA lesions in vivo. Genes [Internet]. 2023 Feb 14;14(2). Available from: http://dx.doi.org/10.3390/genes14020485
154. Ambrosio EMG, Bailey CSL, Unterweger IA, Christensen JB, Bruchez MP, Lundegaard PR, et al. LiverZap: A chemoptogenetic tool for global and locally restricted hepatocyte ablation to study cellular behaviours in liver regeneration. BioRxiv [Internet]. 2023 Mar 14; Available from: http://biorxiv.org/lookup/doi/10.1101/2023.03.13.531762
155. Deng Z, Li L, Jia H, Li NF, He J, Li MD, et al. Insights into the Photodynamics of Fluorescence Emission and Singlet Oxygen Generation of Fluorogen Activating Protein-Malachite Green Systems. Chem Eur J [Internet]. 2023 Mar 16;29(16):e202203684. Available from: http://dx.doi.org/10.1002/chem.202203684
156. Liang P, Zhang Y, Schmidt BF, Ballou B, Qian W, Dong Z, et al. Esterase-Activated, pH-Responsive, and Genetically Targetable Nano-Prodrug for Cancer Cell Photo-Ablation. Small [Internet]. 2023 May [cited 2024 Mar 24];19(19):e2207535. Available from: http://dx.doi.org/10.1002/smll.202207535
157. Bulgari D, Cavolo SL, Schmidt BF, Buchan K, Bruchez MP, Deitcher DL, et al. Ca2+ and cAMP open differentially dilating synaptic fusion pores. J Cell Sci [Internet]. 2023 Jul 136(13). Available from: http://dx.doi.org/10.1242/jcs.261026
158.Weaver N, Hammonds J, Ding L, Lerner G, Dienger-Stambaugh K, Spearman P. KIF16B Mediates Anterograde Transport and Modulates Lysosomal Degradation of the HIV-1 Envelope Glycoprotein. J Virol [Internet]. 2023 Jul 27;97(7):e0025523. Available from: http://dx.doi.org/10.1128/jvi.00255-23
159. Burton AH, Jiao B, Bai Q, Van Laar VS, Wheeler TB, Watkins SC, et al. Full-field exposure of larval zebrafish to narrow waveband LED light sources at defined power and energy for optogenetic applications. J Neurosci Methods [Internet]. 2024;401:110001. Available from: http://dx.doi.org/10.1016/j.jneumeth.2023.110001
160. Prentice JA, van de Weerd R, Bridges AA. Cell-lysis sensing drives biofilm formation in Vibrio cholerae. Nat Commun [Internet]. 2024 Mar 6;15(1):2018. Available from: http://dx.doi.org/10.1038/s41467-024-46399-1